NK012 Fapon NextGen Universal FS DNA Library Prep Kit
Fapon NextGen Library Prep Kits utilize fast, streamlined workflows and novel reagents designed for performance with a broad range of input amounts.
Product Descriptions
The Fapon NextGen Universal FS DNA Library Prep Kit provides a fast, streamlined process for enzymatic fragmentation, end repair and dA tailing in a single tube of enzyme mixture. High-yield, high-quality libraries can be prepared in less than an hour for different input amounts with reduced consumption of consumables. Designed for simplicity and efficiency, the kit utilizes the same process for all DNA input amounts.
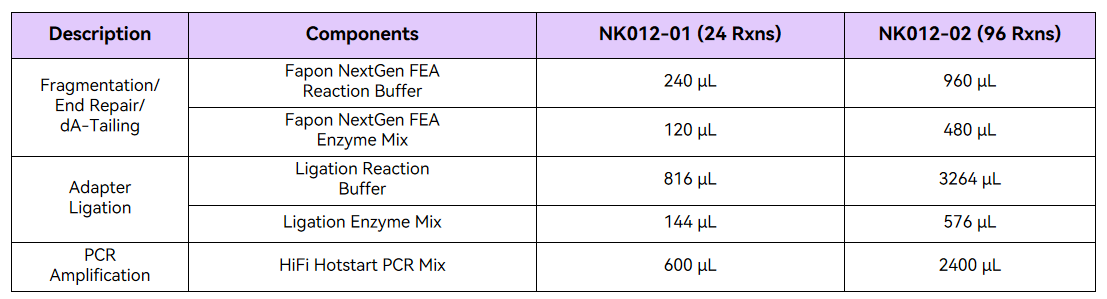
Specifications & Components
Applications
Whole genome sequencing (WGS), including PCR-free
Whole exome sequencing (WES)
Metagenomic analysis
Targeted sequencing
Workflow
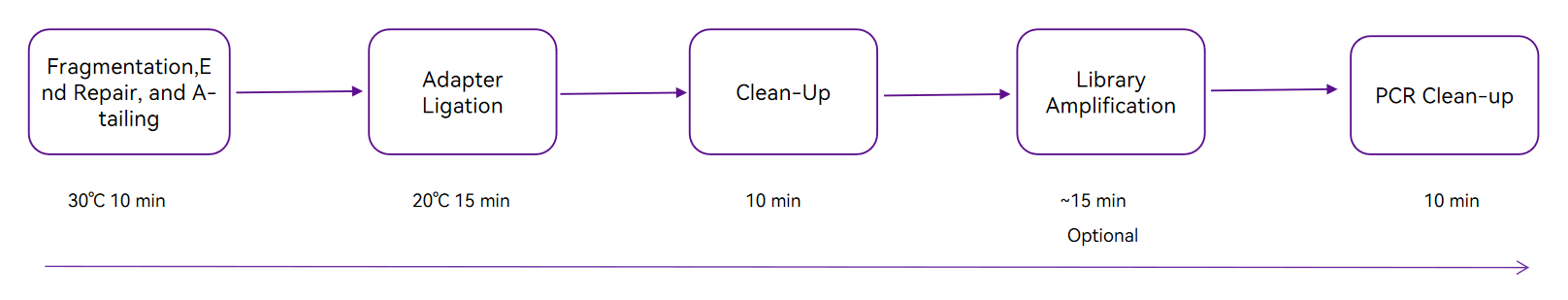
Simplified and easily automated workflow
Total time ~60 minutes
Highlights

Compatible with DNA from different sources (human, animal, plant, microorganism, etc.), and can stably process complex clinical specimens such as saliva, buccal swabs, whole blood, plasma and tumor genomic DNA.
Tunable fragmentation delivers consistent library sizes over a wide input range (1 ng to 500 ng) , and for 0.1 ng to 1ng input amounts, library construction can also be attempted.
Streamlined workflow in less than 1 hour, modular design compatible with automation platforms.
DNA fragmentation, end repair and dA tailing in a single-tube workflow means less sample, plastic and consumable waste
Supports PCR/PCR-Free library construction, and can be adapted to a variety of sequencing applications.
Low background nucleic acid residue and high library conversion rate.
Key Performance Data
Uniformity of Enzymatic Fragmentation
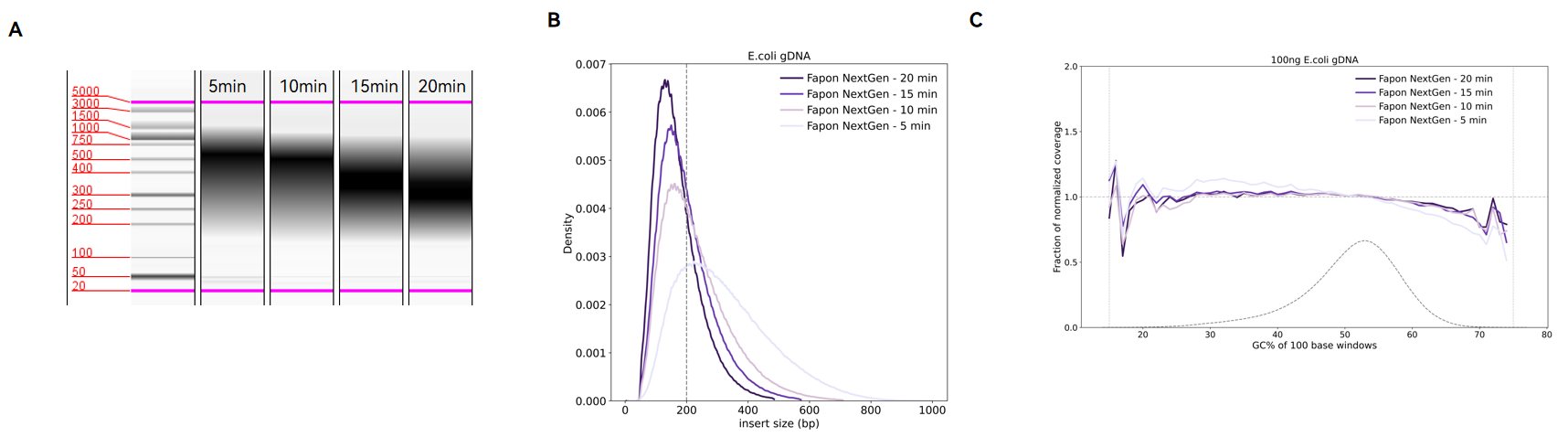
Results of library construction with different fragmentation times.
The library was constructed with 100 ng E. coli DNA as template with different fragmentation time under incubation at 30℃. (A) Library size distribution by Qsep assay. (B) DNA fragment size distribution after DNB-T7 PE 150 bp sequencing. (C) E. coli genome coverage analysis after sequencing.
Superior and Uniform Sequence Coverage
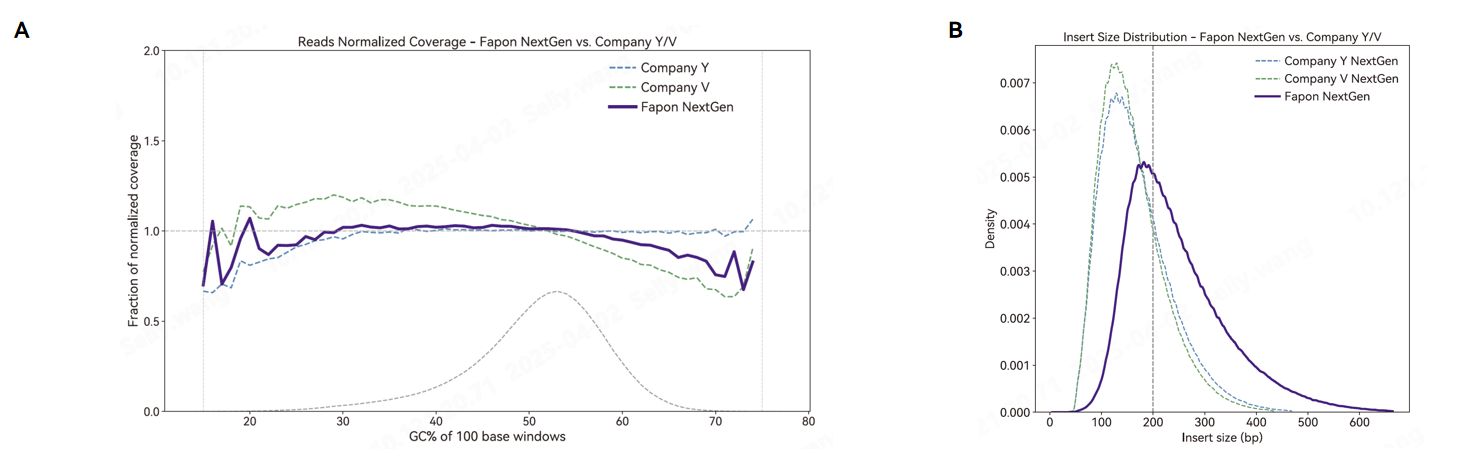
Comparison of the performance of different products for library preparation and sequencing using 100ng E. coli DNA as template.The Reads coverage of Fapon NextGen is more uniform. (A) E. coli genome coverage after sequencing. (B) DNA insert size distribution.
Compatibility with A Range of DNA inputs
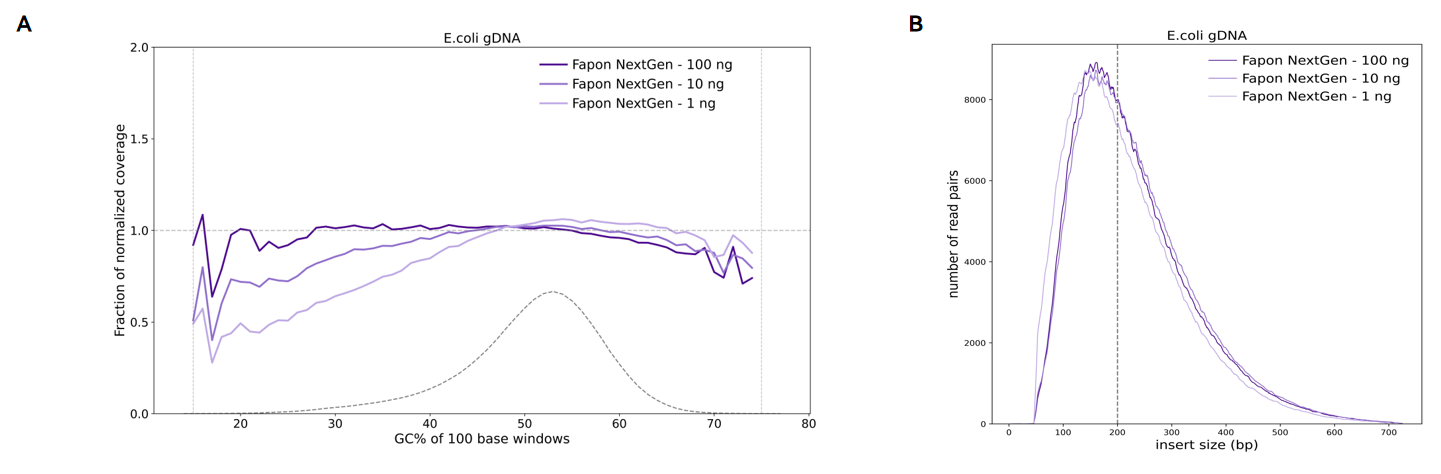
Fapon NextGen Universal FS DNA Library Prep Kit produces libraries with uniform GC coverage and insert size from a range of input amounts.
100ng, 10ng, and 1ng E. coli DNA were used as templates, and the library was constructed by fragmentation at 30°C for 10min. (A) Analysis of E. coli genome coverage after sequencing. Fapon NextGen performed well in terms of Reads coverage at different inputs with 1 ng, 10 ng and 100 ng of E. coli DNA as template.(B)DNA insert sizes across a range of DNA input amounts (1ng - 100 ng). The Reads coverage of Fapon NextGen is more uniform.
Supports PCR/PCR-Free Library Preparation
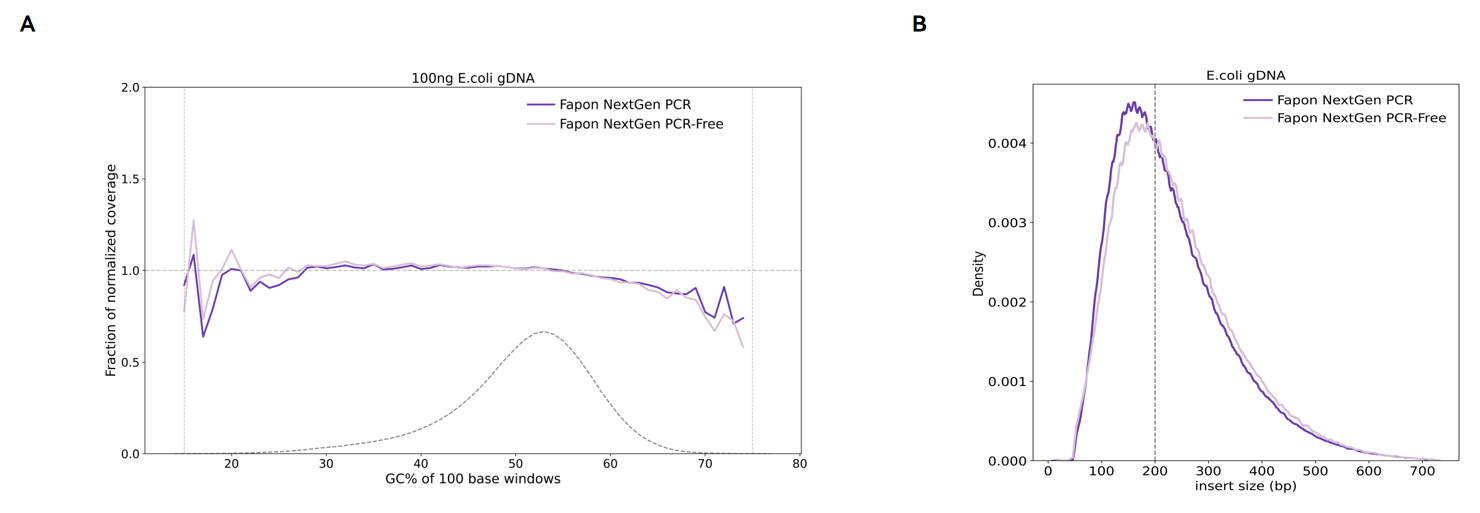
Results of library preparation by PCR/PCR-free.
100 ng E. coli DNA was used as a template and fragmented by incubation at 30°C for 10min without PCR. (A) E. coli genome coverage after sequencing. (B) DNA insert size distribution.
Good Sample Compatibility
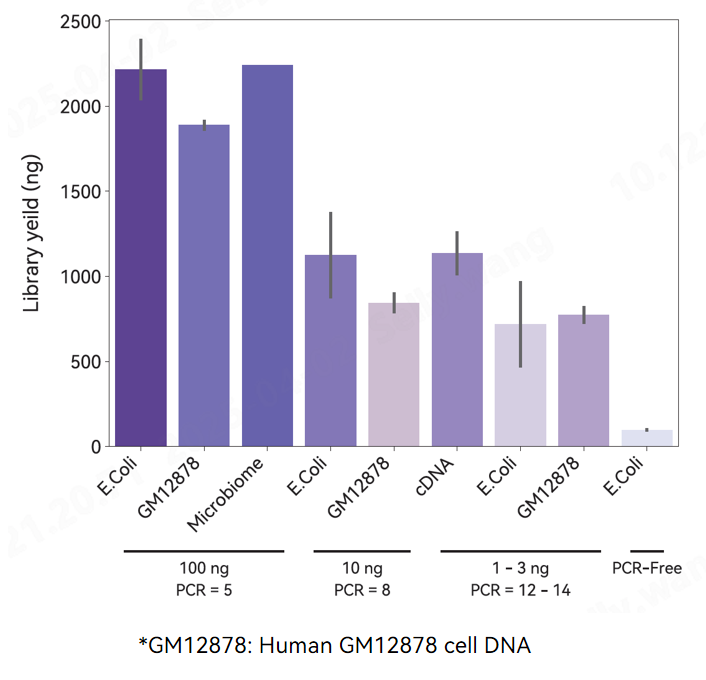
Library yield.
Different PCR cycles were used to achieve the library yield (1ug) requirement using different inputs and sources of DNA as templates, incubated at 30°C 10min for fragmentation.
Precise Mutation Detection Rate
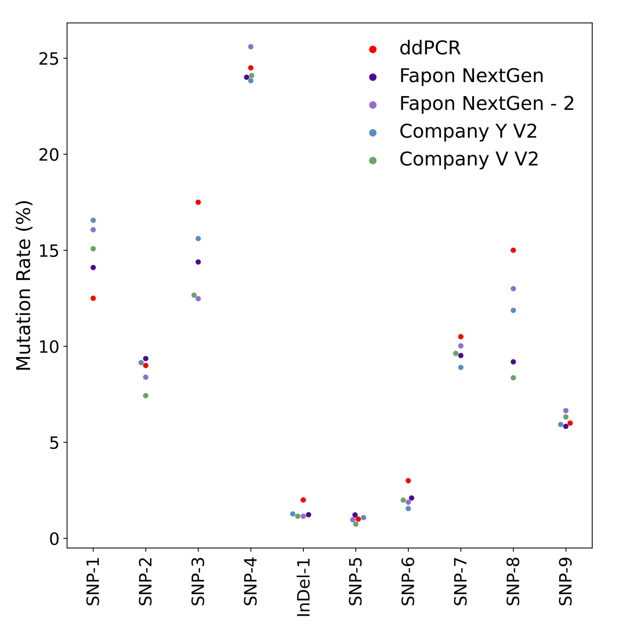
Combined with probe hybridization sequencing, 1-25% mutations in tumor specimens can be accurately detected.
Taking 100ng tumor SNV 1-25% gDNA standard as template, the library was constructed and then hybridized with probe.Fapon NextGen Universal FS DNA Library Prep Kit results are basically consistent with the theoretical values. The mutation detection results are comparable to those of competing products.
-
QA
Join us for a better future of IVD.
If you have any question or need any support, please fill out the information below.